
I recently found notes from my first Supercomputing conference in 2009. First line read “RDBMS is incapable of scaling” and it reminded me of a nice project I worked on in 2010.
Fresh out of Gradschool, I landed at this agricultural company called Genus Plc. I was the “HPC Manager and Scientific Programmer” there, but after parallelizing a few of their production applications the first few months, I realized they also had a (big) data problem. I was about to add Data Engineer to my already long title, before it was cool.
I worked out of one of their subsidiaries, PIC, meaning the Pig Improvement Company. PIC was headquartered near Nashville, TN, and used phenotypic and genotype data to breed seedstock. They weren’t trying to create super pigs, at least not in the sense the media portrays franken-livestock. The goal was to create animals that were more robust and healthier. Simply breeding your “best pigs” the old way can lead to inbreeding and undesirable results.
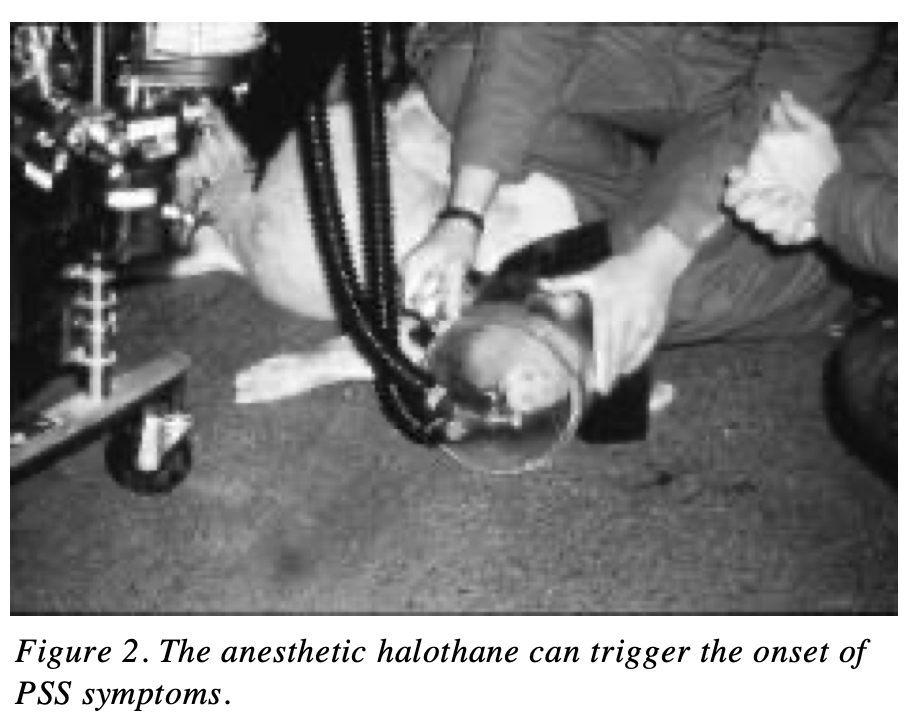
The first application of this approach was used to breed out a gene that caused animals to heat up and die from heart attack. The industry developed a panel to identify this faulty ryanodine receptor gene and started to remove these carriers from the herd (i.e. not breed them). Before this gene was known, the way to test for this condition, know as porcine stress syndrome, was for a vet to give a young pig anesthesia through the snout and listen for an elevated heart rate. If left undiscovered, the animals would die from any stress, even just being transported for example.
PIC collected decades of phenotypic data on their animals and they were accumulating an ever increasing amount of genomic data. The cost of genetic testing had dropped considerably and they went from some 200+ genes on a panel to a whopping 60,000. Deep sequencing data was also starting to come in from vendors like Illumina and BGI. When it was just a few SNPs the SQL database handled the data ok. So as to have as little impact on the business users, a data dump was done more or less weekly, at nights and weekends. But things were getting slower and at 90GBs for a single animal, each data drop was harder and harder to search through.
My knowledge of databases at that time was limited to running a (now defunct) website that died because I botched a database migration to a new host. But PIC was spending a lot of money on consultants to optimize our database, and with naiveté and Dunning-Kruger at my side, I set out to solve this problem. I proposed we move out of the big database and move into a new one just for the research team during a meeting and I’ll never forget the words of geneticist in charge: “You computer people are all the same, you make big promises and leave us with complicated broken systems.” oof, tough crowd.
What they meant was that if it’s not something they can understand and maintain, they rather not be dead in the water after I move on. And this absolutely had come from past experience. Valuable lesson learned, and I pivoted my approach by asking my manager if I could sit in the meetings with the database consultants. I needed to understand the data more, but more importantly I wanted to understand why a relational database was buckling under the weight of what seemed at the time, a reasonable amount of data for it to handle.
But it’s late and to continue with what happened next will require a second post. See you soon and thanks for reading!
Happy Computing Y’all.
Leave a comment